Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK01830
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK01830 |
| Clone name | ej00993 |
| Vector information | |
| Symbol | S1PR3 |
| cDNA sequence | DNA sequence (4180 bp) Predicted protein sequence (399 aa) |
|
HaloTag ORF Clone |
FHC01830

|
| Flexi ORF Clone | FXC01830 |
| Description | Sphingosine 1-phosphate receptor Edg-3 (S1P receptor Edg-3) (Endothelial differentiation G-protein coupled receptor 3) (Sphingosine 1-phosphate receptor 3) (S1P3). |
 Length: 4180 bp
Length: 4180 bp Physical map
Physical map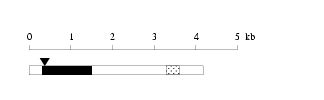
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : YES
Warning for coding interruption : YES Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 2666 bp |
|---|---|
| Genome contig ID | gi89161216f_90696201 |
| PolyA signal sequence (AATAAA,-23) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (113539 - 113588) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 399 aa
Length: 399 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)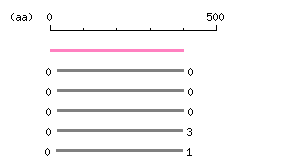 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
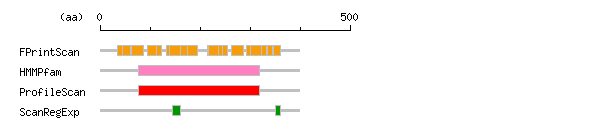
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR004061 | 34 | 46 | PR01523 | Sphingosine 1-phosphate receptor |
| IPR004062 | 45 | 61 | PR01524 | EDG-3 sphingosine 1-phosphate receptor | |
| IPR000276 | 62 | 86 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000276 | 95 | 116 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR004061 | 113 | 122 | PR01523 | Sphingosine 1-phosphate receptor | |
| IPR004061 | 133 | 144 | PR01523 | Sphingosine 1-phosphate receptor | |
| IPR000276 | 139 | 161 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR004062 | 160 | 176 | PR01524 | EDG-3 sphingosine 1-phosphate receptor | |
| IPR000276 | 174 | 195 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR004061 | 215 | 225 | PR01523 | Sphingosine 1-phosphate receptor | |
| IPR000276 | 217 | 240 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR004061 | 236 | 245 | PR01523 | Sphingosine 1-phosphate receptor | |
| IPR004062 | 244 | 254 | PR01524 | EDG-3 sphingosine 1-phosphate receptor | |
| IPR000276 | 262 | 286 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR004061 | 293 | 304 | PR01523 | Sphingosine 1-phosphate receptor | |
| IPR000276 | 301 | 327 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR004061 | 322 | 332 | PR01523 | Sphingosine 1-phosphate receptor | |
| IPR004062 | 335 | 344 | PR01524 | EDG-3 sphingosine 1-phosphate receptor | |
| IPR004062 | 347 | 361 | PR01524 | EDG-3 sphingosine 1-phosphate receptor | |
| HMMPfam | IPR000276 | 77 | 319 | PF00001 | Rhodopsin-like GPCR superfamily |
| ProfileScan | IPR000276 | 77 | 319 | PS50262 | Rhodopsin-like GPCR superfamily |
| ScanRegExp | IPR000276 | 145 | 161 | PS00237 | Rhodopsin-like GPCR superfamily |
| IPR000194 | 351 | 360 | PS00152 | ATPase |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 7 | ALSWILELIVCECQVMATALPPR | 29 | SECONDARY | 23 | 2 | 62 | LTTVLFLVICSFIVLENLMVLIA | 84 | PRIMARY | 23 | 3 | 95 | MYFFIGNLALCDLLAGIAYKVNI | 117 | SECONDARY | 23 | 4 | 138 | SMFVALGASTCSLLAIAIERHLT | 160 | SECONDARY | 23 | 5 | 175 | VFLLIGMCWLIAFTLGALPILGW | 197 | PRIMARY | 23 | 6 | 221 | CISIFTAILVTIVILYARIYFLV | 243 | PRIMARY | 23 | 7 | 263 | LRTVVIVVSVFIACWSPLFILFL | 285 | PRIMARY | 23 | 8 | 298 | LFKAQWFIVLAVLNSAMNPVIYT | 320 | SECONDARY | 23 |
|---|