Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK02116
|
Order Kazusa clone(s) from : |
| Accession No | AB209198 |
|---|---|
| Product ID | ORK02116 |
| Clone name | fj03875 |
| Vector information | |
| Symbol | EDNRB |
| cDNA sequence | DNA sequence (4277 bp) Predicted protein sequence (459 aa) |
|
HaloTag ORF Clone |
FHC02116

|
| Flexi ORF Clone | FXC02116 |
| Description | Endothelin B receptor precursor (ET-B) (Endothelin receptor Non- selective type). |
 Length: 4277 bp
Length: 4277 bp Physical map
Physical map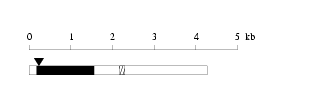
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: NO
Warning for N-terminal truncation: NO Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 2711 bp |
|---|---|
| Genome contig ID | gi51511729r_77267625 |
| PolyA signal sequence (ATTAAA,-21) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (100000 - 99951) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 459 aa
Length: 459 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)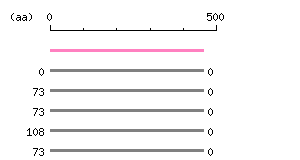 |
|
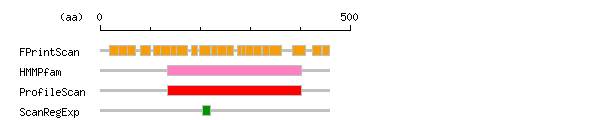
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR001112 | 19 | 37 | PR00571 | Endothelin B receptor |
| IPR001112 | 37 | 54 | PR00571 | Endothelin B receptor | |
| IPR001112 | 55 | 71 | PR00571 | Endothelin B receptor | |
| IPR001112 | 81 | 101 | PR00571 | Endothelin B receptor | |
| IPR000499 | 107 | 126 | PR00366 | Endothelin receptor | |
| IPR000276 | 120 | 144 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000499 | 141 | 153 | PR00366 | Endothelin receptor | |
| IPR000276 | 153 | 174 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR001112 | 183 | 194 | PR00571 | Endothelin B receptor | |
| IPR000276 | 198 | 220 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000499 | 222 | 238 | PR00366 | Endothelin receptor | |
| IPR000276 | 234 | 255 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000499 | 252 | 266 | PR00366 | Endothelin receptor | |
| IPR001112 | 275 | 289 | PR00571 | Endothelin B receptor | |
| IPR000499 | 282 | 300 | PR00366 | Endothelin receptor | |
| IPR000276 | 291 | 314 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000499 | 306 | 323 | PR00366 | Endothelin receptor | |
| IPR000499 | 324 | 338 | PR00366 | Endothelin receptor | |
| IPR000276 | 338 | 362 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000276 | 385 | 411 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR001112 | 425 | 443 | PR00571 | Endothelin B receptor | |
| IPR001112 | 444 | 458 | PR00571 | Endothelin B receptor | |
| HMMPfam | IPR000276 | 135 | 403 | PF00001 | Rhodopsin-like GPCR superfamily |
| ProfileScan | IPR000276 | 135 | 403 | PS50262 | Rhodopsin-like GPCR superfamily |
| ScanRegExp | IPR000276 | 204 | 220 | PS00237 | Rhodopsin-like GPCR superfamily |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 22 | PSLCGRALVALVLACGLSRIWGE | 44 | SECONDARY | 23 | 2 | 117 | FKYINTVVSCLVFVLGIIGNSTL | 139 | PRIMARY | 23 | 3 | 156 | LIASLALGDLLHIVIDIPINVYK | 178 | SECONDARY | 23 | 4 | 191 | CKLVPFIQKASVGITVLSLCALS | 213 | SECONDARY | 23 | 5 | 238 | EIVLIWVVSVVLAVPEAIGFDII | 260 | PRIMARY | 23 | 6 | 294 | LFSFYFCLPLAITAFFYTLMTCE | 316 | SECONDARY | 23 | 7 | 342 | VFCLVLVFALCWLPLHLSRILKL | 364 | PRIMARY | 23 | 8 | 375 | CELLSFLLVLDYIGINMASLNSC | 397 | SECONDARY | 23 |
|---|