Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK11801
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK11801 |
| Clone name | eg01330 |
| Vector information | |
| Symbol | SLIT2 |
| cDNA sequence | DNA sequence (6634 bp) Predicted protein sequence (1562 aa) |
| Description | Slit homolog 2 protein Precursor (Slit-2) [Contains Slit homolog 2 protein N-product;Slit homolog 2 protein C-product] |
 Length: 6634 bp
Length: 6634 bp Physical map
Physical map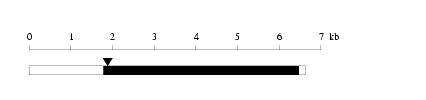
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: NO
Warning for N-terminal truncation: NO Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 157 bp |
|---|---|
| Genome contig ID | gi89161207f_19762650 |
| PolyA signal sequence (AATAAA,-26) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (467239 - 467288) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1562 aa
Length: 1562 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)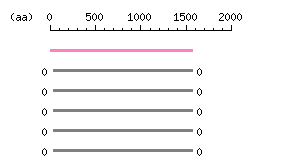 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
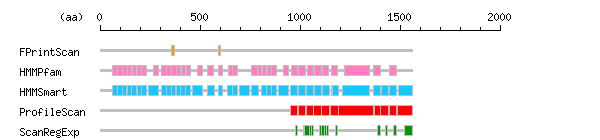
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR001611 | 359 | 372 | PR00019 | Leucine-rich repeat |
| IPR001611 | 590 | 603 | PR00019 | Leucine-rich repeat | |
| HMMPfam | IPR000372 | 60 | 87 | PF01462 | Leucine-rich repeat |
| IPR001611 | 89 | 111 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 113 | 135 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 137 | 157 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 161 | 183 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 185 | 207 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 209 | 231 | PF00560 | Leucine-rich repeat | |
| IPR000483 | 266 | 291 | PF01463 | Cysteine-rich flanking region | |
| IPR000372 | 305 | 332 | PF01462 | Leucine-rich repeat | |
| IPR001611 | 334 | 356 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 358 | 380 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 382 | 404 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 406 | 428 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 430 | 452 | PF00560 | Leucine-rich repeat | |
| IPR000483 | 487 | 512 | PF01463 | Cysteine-rich flanking region | |
| IPR000372 | 538 | 565 | PF01462 | Leucine-rich repeat | |
| IPR001611 | 592 | 614 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 640 | 662 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 664 | 686 | PF00560 | Leucine-rich repeat | |
| IPR000372 | 759 | 786 | PF01462 | Leucine-rich repeat | |
| IPR001611 | 788 | 809 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 811 | 833 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 835 | 857 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 859 | 881 | PF00560 | Leucine-rich repeat | |
| IPR000483 | 916 | 941 | PF01463 | Cysteine-rich flanking region | |
| IPR006209 | 955 | 987 | PF00008 | EGF-like | |
| IPR006209 | 994 | 1028 | PF00008 | EGF-like | |
| IPR006209 | 1035 | 1066 | PF00008 | EGF-like | |
| IPR006209 | 1073 | 1106 | PF00008 | EGF-like | |
| IPR006209 | 1113 | 1144 | PF00008 | EGF-like | |
| IPR006209 | 1158 | 1189 | PF00008 | EGF-like | |
| IPR012680 | 1221 | 1349 | PF02210 | Laminin G | |
| IPR006209 | 1369 | 1400 | PF00008 | EGF-like | |
| IPR006209 | 1449 | 1480 | PF00008 | EGF-like | |
| HMMSmart | IPR000372 | 60 | 92 | SM00013 | Leucine-rich repeat |
| IPR003591 | 86 | 110 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 111 | 134 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 135 | 158 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 159 | 182 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 187 | 206 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 207 | 230 | SM00369 | Leucine-rich repeat | |
| IPR000483 | 242 | 291 | SM00082 | Cysteine-rich flanking region | |
| IPR000372 | 305 | 337 | SM00013 | Leucine-rich repeat | |
| IPR003591 | 331 | 355 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 356 | 379 | SM00369 | Leucine-rich repeat | |
| NULL | 356 | 377 | SM00365 | NULL | |
| IPR003591 | 380 | 403 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 404 | 427 | SM00369 | Leucine-rich repeat | |
| NULL | 428 | 446 | SM00365 | NULL | |
| IPR003591 | 428 | 451 | SM00369 | Leucine-rich repeat | |
| IPR000483 | 463 | 512 | SM00082 | Cysteine-rich flanking region | |
| IPR000372 | 538 | 570 | SM00013 | Leucine-rich repeat | |
| NULL | 590 | 611 | SM00365 | NULL | |
| IPR003591 | 590 | 613 | SM00369 | Leucine-rich repeat | |
| NULL | 638 | 659 | SM00365 | NULL | |
| IPR003591 | 638 | 661 | SM00369 | Leucine-rich repeat | |
| NULL | 662 | 683 | SM00365 | NULL | |
| IPR003591 | 662 | 685 | SM00369 | Leucine-rich repeat | |
| IPR000483 | 697 | 746 | SM00082 | Cysteine-rich flanking region | |
| IPR000372 | 759 | 791 | SM00013 | Leucine-rich repeat | |
| NULL | 809 | 830 | SM00365 | NULL | |
| IPR003591 | 809 | 832 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 833 | 856 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 857 | 880 | SM00369 | Leucine-rich repeat | |
| IPR000483 | 892 | 941 | SM00082 | Cysteine-rich flanking region | |
| IPR001881 | 951 | 988 | SM00179 | EGF-like calcium-binding | |
| IPR006210 | 954 | 988 | SM00181 | EGF | |
| IPR006210 | 993 | 1029 | SM00181 | EGF | |
| IPR001881 | 994 | 1029 | SM00179 | EGF-like calcium-binding | |
| IPR001881 | 1031 | 1067 | SM00179 | EGF-like calcium-binding | |
| IPR003645 | 1034 | 1056 | SM00274 | Follistatin-like | |
| IPR006210 | 1034 | 1067 | SM00181 | EGF | |
| IPR001881 | 1069 | 1107 | SM00179 | EGF-like calcium-binding | |
| IPR006210 | 1072 | 1107 | SM00181 | EGF | |
| IPR001881 | 1109 | 1145 | SM00179 | EGF-like calcium-binding | |
| IPR006210 | 1112 | 1145 | SM00181 | EGF | |
| IPR006210 | 1157 | 1190 | SM00181 | EGF | |
| IPR003645 | 1157 | 1179 | SM00274 | Follistatin-like | |
| IPR001881 | 1160 | 1190 | SM00179 | EGF-like calcium-binding | |
| IPR001791 | 1213 | 1349 | SM00282 | Laminin G | |
| IPR006210 | 1368 | 1401 | SM00181 | EGF | |
| IPR001881 | 1369 | 1401 | SM00179 | EGF-like calcium-binding | |
| IPR006210 | 1407 | 1440 | SM00181 | EGF | |
| IPR003645 | 1448 | 1470 | SM00274 | Follistatin-like | |
| IPR006210 | 1448 | 1481 | SM00181 | EGF | |
| IPR006207 | 1492 | 1561 | SM00041 | Cystine knot | |
| ProfileScan | IPR000742 | 953 | 988 | PS50026 | EGF-like |
| IPR000742 | 990 | 1029 | PS50026 | EGF-like | |
| IPR000742 | 1031 | 1067 | PS50026 | EGF-like | |
| IPR000742 | 1069 | 1107 | PS50026 | EGF-like | |
| IPR000742 | 1109 | 1145 | PS50026 | EGF-like | |
| IPR000742 | 1154 | 1190 | PS50026 | EGF-like | |
| IPR001791 | 1193 | 1366 | PS50025 | Laminin G | |
| IPR000742 | 1370 | 1401 | PS50026 | EGF-like | |
| IPR000742 | 1404 | 1440 | PS50026 | EGF-like | |
| IPR000742 | 1445 | 1481 | PS50026 | EGF-like | |
| IPR006207 | 1486 | 1561 | PS01225 | Cystine knot | |
| ScanRegExp | IPR013032 | 976 | 987 | PS00022 | EGF-like region |
| IPR013032 | 976 | 987 | PS01186 | EGF-like region | |
| IPR013032 | 1017 | 1028 | PS00022 | EGF-like region | |
| IPR013032 | 1017 | 1028 | PS01186 | EGF-like region | |
| IPR001881 | 1024 | 1055 | PS01187 | EGF-like calcium-binding | |
| IPR000152 | 1046 | 1057 | PS00010 | Aspartic acid and asparagine hydroxylation site | |
| IPR013032 | 1055 | 1066 | PS00022 | EGF-like region | |
| IPR013032 | 1095 | 1106 | PS00022 | EGF-like region | |
| IPR013032 | 1095 | 1106 | PS01186 | EGF-like region | |
| IPR001881 | 1109 | 1133 | PS01187 | EGF-like calcium-binding | |
| IPR000152 | 1124 | 1135 | PS00010 | Aspartic acid and asparagine hydroxylation site | |
| IPR013032 | 1133 | 1144 | PS00022 | EGF-like region | |
| IPR013032 | 1133 | 1144 | PS01186 | EGF-like region | |
| IPR013032 | 1178 | 1189 | PS00022 | EGF-like region | |
| IPR013032 | 1178 | 1189 | PS01186 | EGF-like region | |
| IPR013032 | 1389 | 1400 | PS00022 | EGF-like region | |
| IPR013032 | 1389 | 1400 | PS01186 | EGF-like region | |
| IPR013032 | 1428 | 1439 | PS00022 | EGF-like region | |
| IPR013032 | 1469 | 1480 | PS00022 | EGF-like region | |
| IPR013032 | 1469 | 1480 | PS01186 | EGF-like region | |
| IPR006207 | 1524 | 1560 | PS01185 | Cystine knot |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 35 | RGVGWQMLSLSLGLVLAILNKVA | 57 | PRIMARY | 23 |
|---|