Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK00916
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK00916 |
| Clone name | fg01285y2 |
| Vector information | |
| Symbol | KIAA1783 |
| cDNA sequence | DNA sequence (9652 bp) Predicted protein sequence (3096 aa) |
|
HaloTag ORF Clone |
FHC00916

|
| Flexi ORF Clone | FXC00916 |
| Description | CDNA: FLJ22686 fis, clone HSI10987. |
 Length: 9652 bp
Length: 9652 bp Physical map
Physical map
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 361 bp |
|---|---|
| Genome contig ID | gi51511734f_70995734 |
| PolyA signal sequence (AATAAA,-17) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (138788 - 138837) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 3096 aa
Length: 3096 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)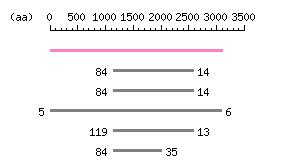 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
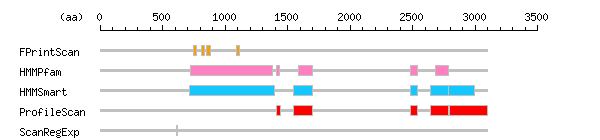
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR001609 | 750 | 769 | PR00193 | Myosin head |
| IPR001609 | 811 | 836 | PR00193 | Myosin head | |
| IPR001609 | 855 | 882 | PR00193 | Myosin head | |
| IPR001609 | 1089 | 1117 | PR00193 | Myosin head | |
| HMMPfam | IPR001609 | 722 | 1382 | PF00063 | Myosin head |
| IPR000048 | 1415 | 1435 | PF00612 | IQ calmodulin-binding region | |
| IPR000857 | 1590 | 1702 | PF00784 | Unconventional myosin/plant kinesin-like protein/non-motor protein conserved region MyTH4 | |
| IPR011511 | 2485 | 2540 | PF07653 | Variant SH3 | |
| IPR000857 | 2683 | 2789 | PF00784 | Unconventional myosin/plant kinesin-like protein/non-motor protein conserved region MyTH4 | |
| HMMSmart | IPR001609 | 714 | 1395 | SM00242 | Myosin head |
| IPR000857 | 1551 | 1702 | SM00139 | Unconventional myosin/plant kinesin-like protein/non-motor protein conserved region MyTH4 | |
| IPR001452 | 2484 | 2541 | SM00326 | Src homology-3 | |
| IPR000857 | 2643 | 2789 | SM00139 | Unconventional myosin/plant kinesin-like protein/non-motor protein conserved region MyTH4 | |
| IPR000299 | 2791 | 2993 | SM00295 | Band 4.1 | |
| ProfileScan | IPR000048 | 1414 | 1443 | PS50096 | IQ calmodulin-binding region |
| IPR000857 | 1551 | 1702 | PS51016 | Unconventional myosin/plant kinesin-like protein/non-motor protein conserved region MyTH4 | |
| IPR001452 | 2481 | 2542 | PS50002 | Src homology-3 | |
| IPR000857 | 2643 | 2789 | PS51016 | Unconventional myosin/plant kinesin-like protein/non-motor protein conserved region MyTH4 | |
| IPR000299 | 2795 | 3096 | PS50057 | Band 4.1 | |
| ScanRegExp | IPR000194 | 613 | 622 | PS00152 | ATPase |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 974 | ELNAVWAVLAAILQLGNICFSSS | 996 | PRIMARY | 23 |
|---|