Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK01096
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK01096 |
| Clone name | hj00601s2 |
| Vector information | |
| Symbol | ARAP2 |
| cDNA sequence | DNA sequence (7580 bp) Predicted protein sequence (1742 aa) |
|
HaloTag ORF Clone |
FHC01096

|
| Flexi ORF Clone | FXC01096 |
| Description | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
 Length: 7580 bp
Length: 7580 bp Physical map
Physical map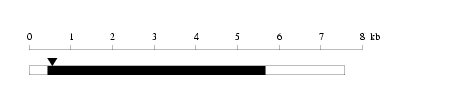
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: NO
Warning for N-terminal truncation: NO Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 1906 bp |
|---|---|
| Genome contig ID | gi89161207r_35644017 |
| PolyA signal sequence (ATTAAA,-27) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (100000 - 99951) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1742 aa
Length: 1742 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)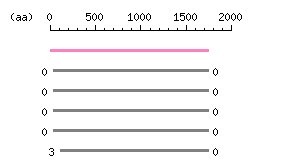 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
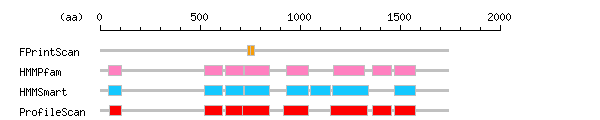
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR001164 | 735 | 754 | PR00405 | Arf GTPase activating protein |
| IPR001164 | 754 | 771 | PR00405 | Arf GTPase activating protein | |
| HMMPfam | IPR001660 | 43 | 106 | PF00536 | Sterile alpha motif SAM |
| IPR001849 | 521 | 612 | PF00169 | Pleckstrin-like | |
| IPR001849 | 626 | 717 | PF00169 | Pleckstrin-like | |
| IPR001164 | 723 | 845 | PF01412 | Arf GTPase activating protein | |
| IPR001849 | 931 | 1041 | PF00169 | Pleckstrin-like | |
| IPR000198 | 1167 | 1320 | PF00620 | RhoGAP | |
| IPR000159 | 1364 | 1458 | PF00788 | Ras-association | |
| IPR001849 | 1473 | 1575 | PF00169 | Pleckstrin-like | |
| HMMSmart | IPR001660 | 42 | 108 | SM00454 | Sterile alpha motif SAM |
| IPR001849 | 521 | 614 | SM00233 | Pleckstrin-like | |
| IPR001849 | 626 | 719 | SM00233 | Pleckstrin-like | |
| IPR001164 | 724 | 845 | SM00105 | Arf GTPase activating protein | |
| IPR001849 | 931 | 1043 | SM00233 | Pleckstrin-like | |
| IPR001849 | 1053 | 1152 | SM00233 | Pleckstrin-like | |
| IPR000198 | 1164 | 1340 | SM00324 | RhoGAP | |
| IPR001849 | 1473 | 1577 | SM00233 | Pleckstrin-like | |
| ProfileScan | IPR001660 | 48 | 108 | PS50105 | Sterile alpha motif SAM |
| IPR001849 | 520 | 612 | PS50003 | Pleckstrin-like | |
| IPR001849 | 625 | 717 | PS50003 | Pleckstrin-like | |
| IPR001164 | 714 | 849 | PS50115 | Arf GTPase activating protein | |
| IPR001849 | 916 | 1041 | PS50003 | Pleckstrin-like | |
| IPR000198 | 1154 | 1335 | PS50238 | RhoGAP | |
| IPR000159 | 1364 | 1458 | PS50200 | Ras-association | |
| IPR001849 | 1472 | 1575 | PS50003 | Pleckstrin-like |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 11 | KPVEKYIFLCVCLIVPLFVTQLS | 33 | PRIMARY | 23 |
|---|