Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK01198
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK01198 |
| Clone name | fh08793 |
| Vector information | |
| Symbol | EPHB6 |
| cDNA sequence | DNA sequence (5166 bp) Predicted protein sequence (1024 aa) |
|
HaloTag ORF Clone |
FHC01198

|
| Flexi ORF Clone | FXC01198 |
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). |
 Length: 5166 bp
Length: 5166 bp Physical map
Physical map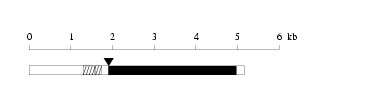
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: NO
Warning for N-terminal truncation: NO Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 188 bp |
|---|---|
| Genome contig ID | gi89161213f_142162166 |
| PolyA signal sequence (ATTAAA,-25) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (116803 - 116852) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1024 aa
Length: 1024 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)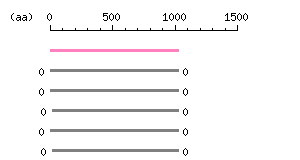 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
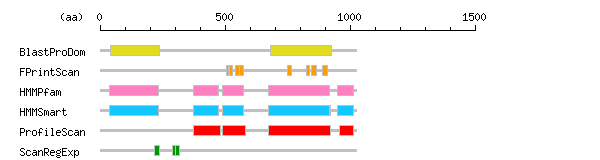
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| BlastProDom | IPR001090 | 43 | 236 | PD001495 | Ephrin receptor |
| IPR000719 | 680 | 925 | PD000001 | Protein kinase | |
| FPrintScan | IPR003962 | 505 | 514 | PR00014 | Fibronectin |
| IPR003962 | 518 | 528 | PR00014 | Fibronectin | |
| IPR003962 | 541 | 559 | PR00014 | Fibronectin | |
| IPR003962 | 559 | 573 | PR00014 | Fibronectin | |
| IPR001245 | 751 | 764 | PR00109 | Tyrosine protein kinase | |
| IPR001245 | 826 | 836 | PR00109 | Tyrosine protein kinase | |
| IPR001245 | 845 | 867 | PR00109 | Tyrosine protein kinase | |
| IPR001245 | 889 | 911 | PR00109 | Tyrosine protein kinase | |
| HMMPfam | IPR001090 | 36 | 235 | PF01404 | Ephrin receptor |
| IPR003961 | 373 | 473 | PF00041 | Fibronectin | |
| IPR003961 | 491 | 575 | PF00041 | Fibronectin | |
| IPR001245 | 673 | 918 | PF07714 | Tyrosine protein kinase | |
| IPR011510 | 948 | 1015 | PF07647 | Sterile alpha motif homology 2 | |
| HMMSmart | IPR001090 | 36 | 235 | SM00615 | Ephrin receptor |
| IPR003961 | 373 | 472 | SM00060 | Fibronectin | |
| IPR003961 | 491 | 572 | SM00060 | Fibronectin | |
| IPR002290 | 673 | 922 | SM00220 | Serine/threonine protein kinase | |
| IPR001245 | 673 | 918 | SM00219 | Tyrosine protein kinase | |
| IPR001660 | 948 | 1015 | SM00454 | Sterile alpha motif SAM | |
| ProfileScan | IPR003961 | 372 | 481 | PS50853 | Fibronectin |
| IPR003961 | 491 | 582 | PS50853 | Fibronectin | |
| IPR000719 | 673 | 922 | PS50011 | Protein kinase | |
| IPR001660 | 959 | 1015 | PS50105 | Sterile alpha motif SAM | |
| ScanRegExp | IPR001426 | 216 | 236 | PS00790 | Receptor tyrosine kinase |
| IPR001426 | 289 | 309 | PS00791 | Receptor tyrosine kinase | |
| IPR013032 | 302 | 317 | PS01186 | EGF-like region |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 20 | VCSLWVLLLVSSVLALEEVLLDT | 42 | PRIMARY | 23 | 2 | 598 | LVIGSILGALAFLLLAAITVLAV | 620 | PRIMARY | 23 |
|---|