Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK01215
|
Order Kazusa clone(s) from : |
| Accession No | AB210004 |
|---|---|
| Product ID | ORK01215 |
| Clone name | fg02848 |
| Vector information | |
| Symbol | NCAN |
| cDNA sequence | DNA sequence (6329 bp) Predicted protein sequence (1335 aa) |
|
HaloTag ORF Clone |
FHC01215

|
| Flexi ORF Clone | FXC01215 |
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). |
 Length: 6329 bp
Length: 6329 bp Physical map
Physical map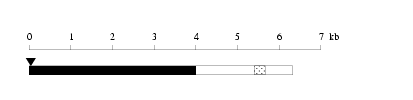
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 2320 bp |
|---|---|
| Genome contig ID | gi42406306f_19088752 |
| PolyA signal sequence (AATAAA,-20) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (135290 - 135339) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1335 aa
Length: 1335 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)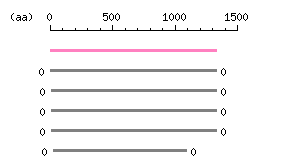 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
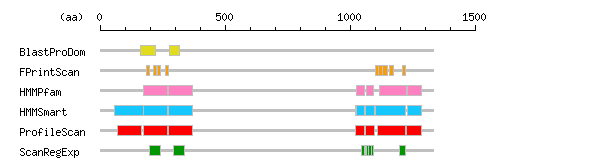
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| BlastProDom | IPR000538 | 163 | 222 | PD000918 | Link |
| IPR000538 | 276 | 319 | PD000918 | Link | |
| FPrintScan | IPR000538 | 184 | 196 | PR01265 | Link |
| IPR000538 | 212 | 225 | PR01265 | Link | |
| IPR000538 | 230 | 241 | PR01265 | Link | |
| IPR000538 | 263 | 272 | PR01265 | Link | |
| IPR002353 | 1101 | 1113 | PR00356 | Type II antifreeze protein | |
| IPR002353 | 1113 | 1130 | PR00356 | Type II antifreeze protein | |
| IPR002353 | 1131 | 1148 | PR00356 | Type II antifreeze protein | |
| IPR002353 | 1158 | 1174 | PR00356 | Type II antifreeze protein | |
| IPR002353 | 1209 | 1222 | PR00356 | Type II antifreeze protein | |
| HMMPfam | IPR000538 | 173 | 268 | PF00193 | Link |
| IPR000538 | 274 | 370 | PF00193 | Link | |
| IPR006209 | 1026 | 1057 | PF00008 | EGF-like | |
| IPR006209 | 1064 | 1095 | PF00008 | EGF-like | |
| IPR001304 | 1119 | 1224 | PF00059 | C-type lectin | |
| IPR000436 | 1229 | 1285 | PF00084 | Sushi/SCR/CCP | |
| HMMSmart | IPR003599 | 58 | 172 | SM00409 | Immunoglobulin subtype |
| IPR000538 | 172 | 269 | SM00445 | Link | |
| IPR000538 | 273 | 371 | SM00445 | Link | |
| IPR001881 | 1022 | 1058 | SM00179 | EGF-like calcium-binding | |
| IPR006210 | 1025 | 1058 | SM00181 | EGF | |
| IPR001881 | 1060 | 1096 | SM00179 | EGF-like calcium-binding | |
| IPR006210 | 1063 | 1096 | SM00181 | EGF | |
| IPR001304 | 1102 | 1223 | SM00034 | C-type lectin | |
| IPR000436 | 1229 | 1285 | SM00032 | Sushi/SCR/CCP | |
| ProfileScan | IPR007110 | 69 | 167 | PS50835 | Immunoglobulin-like |
| IPR000538 | 174 | 269 | PS50963 | Link | |
| IPR000538 | 273 | 371 | PS50963 | Link | |
| IPR000742 | 1022 | 1058 | PS50026 | EGF-like | |
| IPR000742 | 1060 | 1096 | PS50026 | EGF-like | |
| IPR001304 | 1109 | 1223 | PS50041 | C-type lectin | |
| IPR000436 | 1227 | 1287 | PS50923 | Sushi/SCR/CCP | |
| ScanRegExp | IPR000538 | 196 | 241 | PS01241 | Link |
| IPR000538 | 294 | 339 | PS01241 | Link | |
| IPR013032 | 1046 | 1057 | PS00022 | EGF-like region | |
| IPR013032 | 1046 | 1057 | PS01186 | EGF-like region | |
| IPR001881 | 1060 | 1084 | PS01187 | EGF-like calcium-binding | |
| IPR013032 | 1064 | 1075 | PS00022 | EGF-like region | |
| IPR000152 | 1075 | 1086 | PS00010 | Aspartic acid and asparagine hydroxylation site | |
| IPR013032 | 1084 | 1095 | PS00022 | EGF-like region | |
| IPR001304 | 1198 | 1222 | PS00615 | C-type lectin |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 16 | GAPFVWALGLLMLQMLLFVAGEQ | 38 | PRIMARY | 23 | 2 | 58 | SGSVQAALAELVALPCLFTLQPR | 80 | SECONDARY | 23 |
|---|