Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK01285
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK01285 |
| Clone name | ha01215 |
| Vector information | |
| Symbol | ATP2A3 |
| cDNA sequence | DNA sequence (4041 bp) Predicted protein sequence (1043 aa) |
|
HaloTag ORF Clone |
FHC01285

|
| Flexi ORF Clone | FXC01285 |
| Description | ATPase, Ca++ transporting, ubiquitous |
 Length: 4041 bp
Length: 4041 bp Physical map
Physical map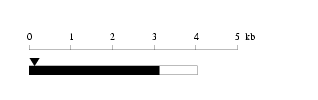
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 908 bp |
|---|---|
| Genome contig ID | gi51511734r_3674557 |
| PolyA signal sequence (None) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (100000 - 99951) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1043 aa
Length: 1043 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)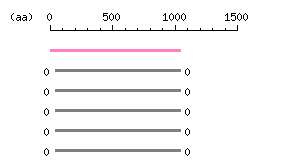 |
|
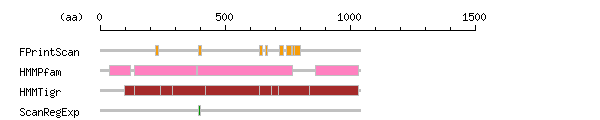
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR001757 | 220 | 234 | PR00119 | ATPase |
| IPR001757 | 393 | 407 | PR00119 | ATPase | |
| IPR001757 | 639 | 650 | PR00119 | ATPase | |
| IPR001757 | 661 | 671 | PR00119 | ATPase | |
| IPR000695 | 717 | 733 | PR00120 | H+ transporting ATPase | |
| IPR000695 | 745 | 761 | PR00120 | H+ transporting ATPase | |
| IPR001757 | 745 | 764 | PR00119 | ATPase | |
| IPR001757 | 768 | 780 | PR00119 | ATPase | |
| IPR000695 | 776 | 801 | PR00120 | H+ transporting ATPase | |
| HMMPfam | IPR004014 | 38 | 121 | PF00690 | ATPase |
| IPR008250 | 137 | 385 | PF00122 | E1-E2 ATPase-associated region | |
| IPR005834 | 389 | 768 | PF00702 | Haloacid dehalogenase-like hydrolase | |
| IPR006068 | 863 | 1034 | PF00689 | ATPase | |
| HMMTigr | IPR005782 | 97 | 1033 | TIGR01116 | Calcium ATPase |
| IPR001757 | 137 | 240 | TIGR01494 | ATPase | |
| IPR001757 | 289 | 420 | TIGR01494 | ATPase | |
| IPR001757 | 636 | 686 | TIGR01494 | ATPase | |
| IPR001757 | 713 | 837 | TIGR01494 | ATPase | |
| ScanRegExp | IPR001757 | 395 | 401 | PS00154 | ATPase |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 102 | EDLLVRILLLAALVSFVLAWFE | 123 | PRIMARY | 22 | 2 | 130 | TAFVEPLVIMLILVANAIVGVWQ | 152 | PRIMARY | 23 | 3 | 302 | RQLSHAISVICVAVWVINIGHFA | 324 | PRIMARY | 23 | 4 | 343 | AVALAVAAIPEGLPAVITTCLAL | 365 | PRIMARY | 23 | 5 | 820 | FLTAILGLPEALIPVQLLWVNLV | 842 | SECONDARY | 23 | 6 | 875 | GWLFFRYLAIGVYVGLATVAAAT | 897 | PRIMARY | 23 | 7 | 939 | FPTTMALSVLVTIEMCNALNSVS | 961 | SECONDARY | 23 | 8 | 977 | LLVAVAMSMALHFLILLVPPLPL | 999 | PRIMARY | 23 | 9 | 1010 | QWVVVLQISLPVILLDEALKYLS | 1032 | PRIMARY | 23 |
|---|