Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK01303
|
Order Kazusa clone(s) from : |
| Accession No | |
|---|---|
| Product ID | ORK01303 |
| Clone name | hk10385 |
| Vector information | |
| Symbol | ATP1A1 |
| cDNA sequence | DNA sequence (3650 bp) Predicted protein sequence (1038 aa) |
|
HaloTag ORF Clone |
FHC01303

|
| Flexi ORF Clone | FXC01303 |
| Description | Sodium/potassium-transporting ATPase subunit alpha-1 precursor (EC 3.6.3.9) (Sodium pump subunit alpha-1) (Na(+)/K(+) ATPase alpha-1 subunit). |
 Length: 3650 bp
Length: 3650 bp Physical map
Physical map
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: NO
Warning for N-terminal truncation: NO Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 328 bp |
|---|---|
| Genome contig ID | gi89161185f_116617407 |
| PolyA signal sequence (AATAAA,-23) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (131512 - 131561) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1038 aa
Length: 1038 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)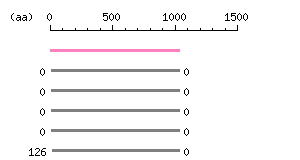 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
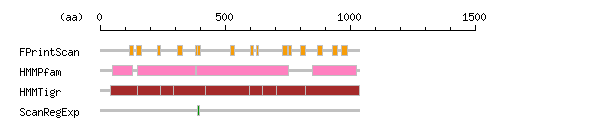
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR006069 | 118 | 132 | PR00121 | ATPase |
| IPR006069 | 145 | 165 | PR00121 | ATPase | |
| IPR001757 | 229 | 243 | PR00119 | ATPase | |
| IPR006069 | 309 | 331 | PR00121 | ATPase | |
| IPR006069 | 382 | 403 | PR00121 | ATPase | |
| IPR001757 | 389 | 403 | PR00119 | ATPase | |
| IPR006069 | 520 | 538 | PR00121 | ATPase | |
| IPR001757 | 602 | 613 | PR00119 | ATPase | |
| IPR001757 | 624 | 634 | PR00119 | ATPase | |
| IPR001757 | 730 | 749 | PR00119 | ATPase | |
| IPR001757 | 754 | 766 | PR00119 | ATPase | |
| IPR006069 | 802 | 823 | PR00121 | ATPase | |
| IPR006069 | 869 | 889 | PR00121 | ATPase | |
| IPR006069 | 931 | 951 | PR00121 | ATPase | |
| IPR006069 | 965 | 989 | PR00121 | ATPase | |
| HMMPfam | IPR004014 | 48 | 131 | PF00690 | ATPase |
| IPR008250 | 150 | 381 | PF00122 | E1-E2 ATPase-associated region | |
| IPR005834 | 385 | 753 | PF00702 | Haloacid dehalogenase-like hydrolase | |
| IPR006068 | 849 | 1027 | PF00689 | ATPase | |
| HMMTigr | IPR005775 | 42 | 1038 | TIGR01106 | ATPase |
| IPR001757 | 150 | 242 | TIGR01494 | ATPase | |
| IPR001757 | 294 | 420 | TIGR01494 | ATPase | |
| IPR001757 | 599 | 649 | TIGR01494 | ATPase | |
| IPR001757 | 704 | 823 | TIGR01494 | ATPase | |
| ScanRegExp | IPR001757 | 391 | 397 | PS00154 | ATPase |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 113 | GGFSMLLWIGAILCFLAYSIQAA | 135 | PRIMARY | 23 | 2 | 145 | LYLGVVLSAVVIITGCFSYYQ | 165 | PRIMARY | 21 | 3 | 307 | IHIITGVAVFLGVSFFILSLILE | 329 | PRIMARY | 23 | 4 | 339 | LIGIIVANVPEGLLATVTVCLTL | 361 | PRIMARY | 23 | 5 | 810 | IANIPLPLGTVTILCIDLGTDMV | 832 | SECONDARY | 23 | 6 | 874 | MIQALGGFFTYFVILAENGFLPI | 896 | SECONDARY | 23 | 7 | 930 | EFTCHTAFFVSIVVVQWADLVIC | 952 | PRIMARY | 23 | 8 | 968 | ILIFGLFEETALAAFLSYCPGMG | 990 | SECONDARY | 23 | 9 | 998 | LKPTWWFCAFPYSLLIFVYDEV | 1019 | SECONDARY | 22 |
|---|