Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK04225
|
Order Kazusa clone(s) from : |
| Accession No | AB209687 |
|---|---|
| Product ID | ORK04225 |
| Clone name | pf08939 |
| Vector information | |
| Symbol | ATP8A1 |
| cDNA sequence | DNA sequence (8074 bp) Predicted protein sequence (1177 aa) |
|
Flexi ORF Clone |
FXC04225

|
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). |
 Length: 8074 bp
Length: 8074 bp Physical map
Physical map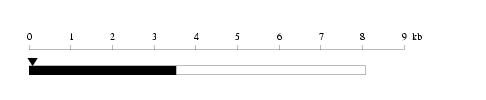
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 4539 bp |
|---|---|
| Genome contig ID | gi89161207r_42005151 |
| PolyA signal sequence (AATAAA,-20) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (100000 - 99951) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1177 aa
Length: 1177 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)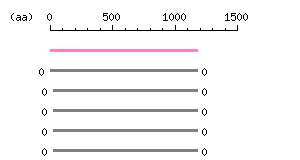 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
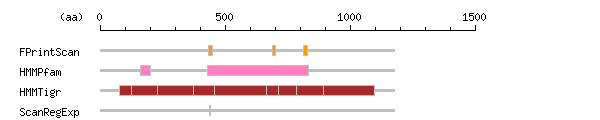
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR001757 | 435 | 449 | PR00119 | ATPase |
| IPR001757 | 690 | 700 | PR00119 | ATPase | |
| IPR001757 | 812 | 831 | PR00119 | ATPase | |
| HMMPfam | IPR008250 | 160 | 203 | PF00122 | E1-E2 ATPase-associated region |
| IPR005834 | 431 | 835 | PF00702 | Haloacid dehalogenase-like hydrolase | |
| HMMTigr | IPR006539 | 77 | 1099 | TIGR01652 | Phospholipid-translocating P-type ATPase |
| IPR001757 | 127 | 229 | TIGR01494 | ATPase | |
| IPR001757 | 372 | 456 | TIGR01494 | ATPase | |
| IPR001757 | 665 | 715 | TIGR01494 | ATPase | |
| IPR001757 | 785 | 895 | TIGR01494 | ATPase | |
| ScanRegExp | IPR001757 | 437 | 443 | PS00154 | ATPase |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 104 | AANSFFLFIALLQQIPDVSPTGR | 126 | SECONDARY | 23 | 2 | 128 | TTLVPLLFILAVAAIKEIIEDI | 149 | PRIMARY | 22 | 3 | 323 | NVQILILFCILIAMSLVCSVGSA | 345 | PRIMARY | 23 | 4 | 370 | LNFLTFIILFNNLIPISLLVTLE | 392 | PRIMARY | 23 | 5 | 772 | QYFLDLALSCKAVICCRVSPLQK | 794 | SECONDARY | 23 | 6 | 869 | SKCILYCFYKNIVLYIIEIWFAF | 891 | SECONDARY | 23 | 7 | 900 | LFERWCIGLYNVMFTAMPPLTLG | 922 | SECONDARY | 23 | 8 | 954 | FWVHCLNGLFHSVILFWFPLKAL | 976 | PRIMARY | 23 | 9 | 989 | DYLLLGNFVYTFVVITVCLKAGL | 1011 | SECONDARY | 23 | 10 | 1019 | FSHIAIWGSIALWVVFFGIYSSL | 1041 | PRIMARY | 23 | 11 | 1058 | LFSSGVFWMGLLFIPVASLLLDV | 1080 | PRIMARY | 23 |
|---|