Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK05495
|
Order Kazusa clone(s) from : |
| Accession No | AB209668 |
|---|---|
| Product ID | ORK05495 |
| Clone name | pf02494 |
| Vector information | |
| Symbol | IGF2R |
| cDNA sequence | DNA sequence (7662 bp) Predicted protein sequence (2414 aa) |
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate |
 Length: 7662 bp
Length: 7662 bp Physical map
Physical map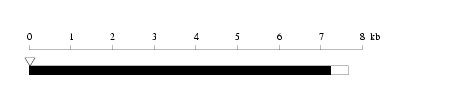
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 415 bp |
|---|---|
| Genome contig ID | gi89161210f_160232286 |
| PolyA signal sequence (None) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (214237 - 214286) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 2414 aa
Length: 2414 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)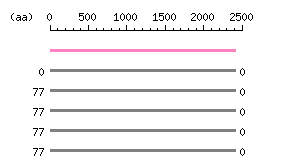 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
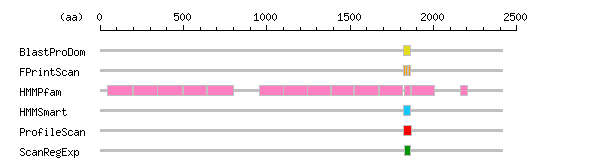
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| BlastProDom | IPR000562 | 1820 | 1865 | PD000995 | Type II fibronectin |
| FPrintScan | IPR000562 | 1823 | 1832 | PR00013 | Type II fibronectin |
| IPR000562 | 1834 | 1846 | PR00013 | Type II fibronectin | |
| IPR000562 | 1849 | 1864 | PR00013 | Type II fibronectin | |
| HMMPfam | IPR000479 | 46 | 194 | PF00878 | Cation-independent mannose-6-phosphate receptor |
| IPR000479 | 200 | 344 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 346 | 496 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 498 | 639 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 643 | 799 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 954 | 1101 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 1102 | 1243 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 1245 | 1385 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 1388 | 1522 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 1526 | 1671 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 1678 | 1817 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000562 | 1826 | 1865 | PF00040 | Type II fibronectin | |
| IPR000479 | 1869 | 2006 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| IPR000479 | 2160 | 2202 | PF00878 | Cation-independent mannose-6-phosphate receptor | |
| HMMSmart | IPR000562 | 1819 | 1865 | SM00059 | Type II fibronectin |
| ProfileScan | IPR000562 | 1821 | 1867 | PS51092 | Type II fibronectin |
| ScanRegExp | IPR000562 | 1826 | 1865 | PS00023 | Type II fibronectin |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 2186 | TADCQYLFSWYTSAVCPLGVGF | 2207 | SECONDARY | 22 | 2 | 2228 | AVGAVLSLLLVALTCCLLALLLY | 2250 | PRIMARY | 23 |
|---|