Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK06714
|
Order Kazusa clone(s) from : |
| Accession No | AB208943 |
|---|---|
| Product ID | ORK06714 |
| Clone name | fk13921 |
| Vector information | |
| Symbol | RXFP1 |
| cDNA sequence | DNA sequence (3671 bp) Predicted protein sequence (627 aa) |
| Description | Relaxin receptor 1 (Relaxin family peptide receptor 1) (Leucine-rich repeat-containing G-protein coupled receptor 7). |
 Length: 3671 bp
Length: 3671 bp Physical map
Physical map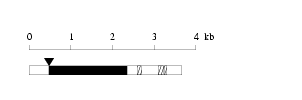
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: NO
Warning for N-terminal truncation: NO Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 1312 bp |
|---|---|
| Genome contig ID | gi89161207f_159562578 |
| PolyA signal sequence (AATAAA,-26) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (231393 - 231442) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 627 aa
Length: 627 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)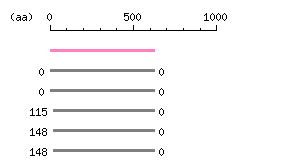 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
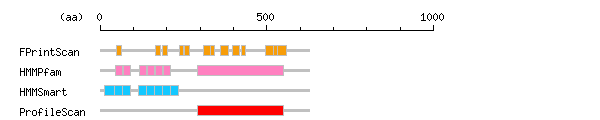
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR008112 | 49 | 64 | PR01739 | Relaxin receptor |
| IPR001611 | 167 | 180 | PR00019 | Leucine-rich repeat | |
| IPR001611 | 188 | 201 | PR00019 | Leucine-rich repeat | |
| IPR008112 | 238 | 250 | PR01739 | Relaxin receptor | |
| IPR008112 | 254 | 268 | PR01739 | Relaxin receptor | |
| IPR000276 | 310 | 331 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR008112 | 332 | 342 | PR01739 | Relaxin receptor | |
| IPR000276 | 362 | 384 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR000276 | 397 | 418 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR008112 | 423 | 436 | PR01739 | Relaxin receptor | |
| IPR000276 | 496 | 520 | PR00237 | Rhodopsin-like GPCR superfamily | |
| IPR008112 | 520 | 534 | PR01739 | Relaxin receptor | |
| IPR000276 | 533 | 559 | PR00237 | Rhodopsin-like GPCR superfamily | |
| HMMPfam | IPR001611 | 45 | 67 | PF00560 | Leucine-rich repeat |
| IPR001611 | 69 | 91 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 118 | 140 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 142 | 164 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 166 | 188 | PF00560 | Leucine-rich repeat | |
| IPR001611 | 190 | 212 | PF00560 | Leucine-rich repeat | |
| IPR000276 | 292 | 551 | PF00001 | Rhodopsin-like GPCR superfamily | |
| HMMSmart | IPR003591 | 14 | 42 | SM00369 | Leucine-rich repeat |
| NULL | 43 | 69 | SM00365 | NULL | |
| IPR003591 | 43 | 66 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 67 | 90 | SM00369 | Leucine-rich repeat | |
| NULL | 116 | 137 | SM00365 | NULL | |
| IPR003591 | 116 | 139 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 140 | 163 | SM00369 | Leucine-rich repeat | |
| NULL | 164 | 182 | SM00365 | NULL | |
| IPR003591 | 164 | 187 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 188 | 211 | SM00369 | Leucine-rich repeat | |
| IPR003591 | 212 | 235 | SM00369 | Leucine-rich repeat | |
| ProfileScan | IPR000276 | 292 | 551 | PS50262 | Rhodopsin-like GPCR superfamily |
 Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments| Method | No. | N terminal | transmembrane region | C terminal | type | length | SOSUI2 | 1 | 274 | VSIIQRVFVWVVSAVTCFGNIFV | 296 | PRIMARY | 23 | 2 | 312 | MSIISLCCADCLMGIYLFVIGGF | 334 | PRIMARY | 23 | 3 | 363 | ILSTEVSVLLLTFLTLEKYICIV | 385 | PRIMARY | 23 | 4 | 398 | TITVLILIWITGFIVAFIPLSNK | 420 | PRIMARY | 23 | 5 | 446 | AQIYSVAIFLGINLAAFIIIVFS | 468 | PRIMARY | 23 | 6 | 500 | FFFIVFTDALCWIPIFVVKFLSL | 522 | PRIMARY | 23 | 7 | 533 | SWVVIFILPINSALNPILYTLTT | 555 | SECONDARY | 23 |
|---|