Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK07173
|
Order Kazusa clone(s) from : |
| Accession No | AB209339 |
|---|---|
| Product ID | ORK07173 |
| Clone name | fh12505 |
| Vector information | |
| Symbol | TNKS |
| cDNA sequence | DNA sequence (5326 bp) Predicted protein sequence (1055 aa) |
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). |
 Length: 5326 bp
Length: 5326 bp Physical map
Physical map
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 2157 bp |
|---|---|
| Genome contig ID | gi51511724f_9375221 |
| PolyA signal sequence (ATTAAA,-23) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (298595 - 298644) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 1055 aa
Length: 1055 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)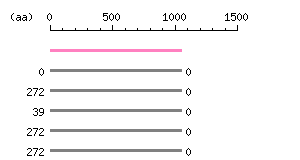 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
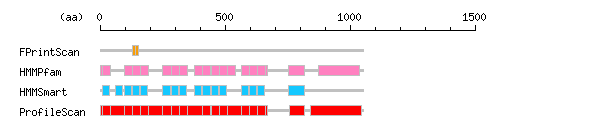
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR002110 | 130 | 142 | PR01415 | Ankyrin |
| IPR002110 | 142 | 154 | PR01415 | Ankyrin | |
| HMMPfam | IPR002110 | 1 | 8 | PF00023 | Ankyrin |
| IPR002110 | 9 | 41 | PF00023 | Ankyrin | |
| IPR002110 | 96 | 128 | PF00023 | Ankyrin | |
| IPR002110 | 129 | 161 | PF00023 | Ankyrin | |
| IPR002110 | 162 | 194 | PF00023 | Ankyrin | |
| IPR002110 | 249 | 284 | PF00023 | Ankyrin | |
| IPR002110 | 285 | 317 | PF00023 | Ankyrin | |
| IPR002110 | 318 | 350 | PF00023 | Ankyrin | |
| IPR002110 | 377 | 408 | PF00023 | Ankyrin | |
| IPR002110 | 411 | 443 | PF00023 | Ankyrin | |
| IPR002110 | 444 | 476 | PF00023 | Ankyrin | |
| IPR002110 | 477 | 509 | PF00023 | Ankyrin | |
| IPR002110 | 510 | 541 | PF00023 | Ankyrin | |
| IPR002110 | 564 | 596 | PF00023 | Ankyrin | |
| IPR002110 | 597 | 629 | PF00023 | Ankyrin | |
| IPR002110 | 630 | 662 | PF00023 | Ankyrin | |
| IPR002110 | 663 | 671 | PF00023 | Ankyrin | |
| IPR011510 | 752 | 817 | PF07647 | Sterile alpha motif homology 2 | |
| IPR001290 | 872 | 1038 | PF00644 | Poly(ADP-ribose) polymerase | |
| HMMSmart | IPR002110 | 9 | 38 | SM00248 | Ankyrin |
| IPR002110 | 62 | 90 | SM00248 | Ankyrin | |
| IPR002110 | 96 | 125 | SM00248 | Ankyrin | |
| IPR002110 | 129 | 158 | SM00248 | Ankyrin | |
| IPR002110 | 162 | 191 | SM00248 | Ankyrin | |
| IPR002110 | 249 | 281 | SM00248 | Ankyrin | |
| IPR002110 | 285 | 314 | SM00248 | Ankyrin | |
| IPR002110 | 318 | 347 | SM00248 | Ankyrin | |
| IPR002110 | 377 | 405 | SM00248 | Ankyrin | |
| IPR002110 | 411 | 440 | SM00248 | Ankyrin | |
| IPR002110 | 444 | 473 | SM00248 | Ankyrin | |
| IPR002110 | 477 | 506 | SM00248 | Ankyrin | |
| IPR002110 | 564 | 593 | SM00248 | Ankyrin | |
| IPR002110 | 597 | 626 | SM00248 | Ankyrin | |
| IPR002110 | 630 | 659 | SM00248 | Ankyrin | |
| IPR001660 | 752 | 817 | SM00454 | Sterile alpha motif SAM | |
| ProfileScan | IPR002110 | 1 | 671 | PS50297 | Ankyrin |
| IPR002110 | 9 | 41 | PS50088 | Ankyrin | |
| IPR002110 | 96 | 128 | PS50088 | Ankyrin | |
| IPR002110 | 129 | 161 | PS50088 | Ankyrin | |
| IPR002110 | 162 | 194 | PS50088 | Ankyrin | |
| IPR002110 | 249 | 284 | PS50088 | Ankyrin | |
| IPR002110 | 285 | 317 | PS50088 | Ankyrin | |
| IPR002110 | 318 | 350 | PS50088 | Ankyrin | |
| IPR002110 | 411 | 443 | PS50088 | Ankyrin | |
| IPR002110 | 444 | 476 | PS50088 | Ankyrin | |
| IPR002110 | 477 | 509 | PS50088 | Ankyrin | |
| IPR002110 | 564 | 596 | PS50088 | Ankyrin | |
| IPR002110 | 597 | 629 | PS50088 | Ankyrin | |
| IPR002110 | 630 | 662 | PS50088 | Ankyrin | |
| IPR001660 | 758 | 817 | PS50105 | Sterile alpha motif SAM | |
| IPR012317 | 840 | 1045 | PS51059 | PARP |