Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK07179
|
Order Kazusa clone(s) from : |
| Accession No | AB209238 |
|---|---|
| Product ID | ORK07179 |
| Clone name | fj15762 |
| Vector information | |
| Symbol | TNS1 |
| cDNA sequence | DNA sequence (4292 bp) Predicted protein sequence (873 aa) |
| Description | Tensin-1. |
 Length: 4292 bp
Length: 4292 bp Physical map
Physical map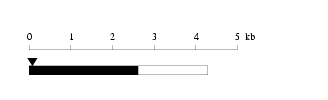
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 1670 bp |
|---|---|
| Genome contig ID | gi89161199r_218275757 |
| PolyA signal sequence (AATAAA,-21) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (100000 - 99951) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 873 aa
Length: 873 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)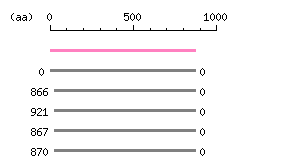 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
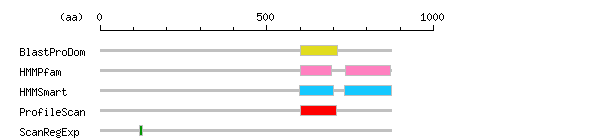
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| BlastProDom | IPR000980 | 601 | 713 | PD000093 | SH2 motif |
| HMMPfam | IPR000980 | 601 | 695 | PF00017 | SH2 motif |
| IPR013625 | 736 | 872 | PF08416 | Tensin phosphotyrosine-binding domain | |
| HMMSmart | IPR000980 | 599 | 701 | SM00252 | SH2 motif |
| IPR006020 | 732 | 873 | SM00462 | Phosphotyrosine interaction region | |
| ProfileScan | IPR000980 | 601 | 710 | PS50001 | SH2 motif |
| ScanRegExp | IPR000209 | 117 | 127 | PS00138 | Peptidase S8 and S53 |