Kazusa Clone (Original Type)
Gene/Protein Characteristic Table for
ORK06529
|
Order Kazusa clone(s) from : |
| Accession No | AB208904 |
|---|---|
| Product ID | ORK06529 |
| Clone name | pf00494 |
| Vector information | |
| Symbol | PTPN13 |
| cDNA sequence | DNA sequence (7911 bp) Predicted protein sequence (2434 aa) |
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). |
 Length: 7911 bp
Length: 7911 bp Physical map
Physical map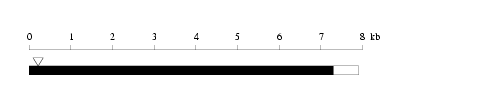
 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq.
Prediction of protein coding region (GeneMark analysis) for :
cloned DNA seq. Warning for N-terminal truncation: YES
Warning for N-terminal truncation: YES Warning for coding interruption : NO
Warning for coding interruption : NO Integrity of 3' end
Integrity of 3' end
| Length of 3'UTR | 606 bp |
|---|---|
| Genome contig ID | gi89161207f_87712540 |
| PolyA signal sequence (AATAAA,-32) |
+----*----+----*----+----*----+---- |
| Flanking genome sequence (242796 - 242845) |
----+----*----+----*----+----*----+----*----+----* |
 Length: 2434 aa
Length: 2434 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)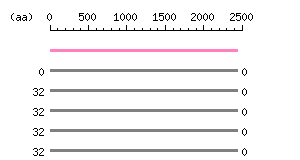 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
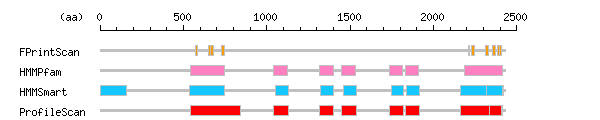
 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI) Result of InterProScan
Result of InterProScan
| Search method | interpro_ID | From | To | Entry | Definition |
|---|---|---|---|---|---|
| FPrintScan | IPR000299 | 573 | 585 | PR00935 | Band 4.1 |
| IPR000299 | 649 | 662 | PR00935 | Band 4.1 | |
| IPR000299 | 662 | 682 | PR00935 | Band 4.1 | |
| IPR000299 | 729 | 745 | PR00935 | Band 4.1 | |
| IPR000242 | 2210 | 2217 | PR00700 | Protein-tyrosine phosphatase | |
| IPR000242 | 2228 | 2248 | PR00700 | Protein-tyrosine phosphatase | |
| IPR000242 | 2315 | 2332 | PR00700 | Protein-tyrosine phosphatase | |
| IPR000242 | 2352 | 2370 | PR00700 | Protein-tyrosine phosphatase | |
| IPR000242 | 2383 | 2398 | PR00700 | Protein-tyrosine phosphatase | |
| IPR000242 | 2399 | 2409 | PR00700 | Protein-tyrosine phosphatase | |
| HMMPfam | IPR000299 | 542 | 749 | PF00373 | Band 4.1 |
| IPR001478 | 1042 | 1125 | PF00595 | PDZ/DHR/GLGF | |
| IPR001478 | 1317 | 1399 | PF00595 | PDZ/DHR/GLGF | |
| IPR001478 | 1450 | 1535 | PF00595 | PDZ/DHR/GLGF | |
| IPR001478 | 1737 | 1815 | PF00595 | PDZ/DHR/GLGF | |
| IPR001478 | 1832 | 1912 | PF00595 | PDZ/DHR/GLGF | |
| IPR000242 | 2186 | 2415 | PF00102 | Protein-tyrosine phosphatase | |
| HMMSmart | IPR011019 | 1 | 158 | SM00750 | KIND |
| IPR000299 | 536 | 749 | SM00295 | Band 4.1 | |
| IPR001478 | 1051 | 1128 | SM00228 | PDZ/DHR/GLGF | |
| IPR001478 | 1325 | 1402 | SM00228 | PDZ/DHR/GLGF | |
| IPR001478 | 1458 | 1538 | SM00228 | PDZ/DHR/GLGF | |
| IPR001478 | 1746 | 1818 | SM00228 | PDZ/DHR/GLGF | |
| IPR001478 | 1840 | 1915 | SM00228 | PDZ/DHR/GLGF | |
| IPR000242 | 2161 | 2418 | SM00194 | Protein-tyrosine phosphatase | |
| IPR003595 | 2316 | 2415 | SM00404 | Protein-tyrosine phosphatase | |
| ProfileScan | IPR000299 | 540 | 840 | PS50057 | Band 4.1 |
| IPR001478 | 1042 | 1128 | PS50106 | PDZ/DHR/GLGF | |
| IPR001478 | 1317 | 1402 | PS50106 | PDZ/DHR/GLGF | |
| IPR001478 | 1450 | 1538 | PS50106 | PDZ/DHR/GLGF | |
| IPR001478 | 1737 | 1818 | PS50106 | PDZ/DHR/GLGF | |
| IPR001478 | 1832 | 1915 | PS50106 | PDZ/DHR/GLGF | |
| IPR000242 | 2162 | 2416 | PS50055 | Protein-tyrosine phosphatase | |
| IPR000387 | 2338 | 2407 | PS50056 | Protein-tyrosine phosphatase |